13 Oct 2014
https://www.ibiology.org/biochemistry/rna-structure/#part-3
Lecture Overview:
In Part 1, Dr. Pyle explains that many RNA molecules have elaborate structures that are essential for their functions. Even mRNA, a relatively linear molecule, can contain distinctive three- dimensional structures. RNA duplexes are the units of secondary structure, and these form in regions where base-pairing occurs. Duplex regions often include internal or terminal loops, and they can contain unusual types of base-pairing. These secondary structural elements can arrange themselves to form highly complex tertiary structures. It is the variety of these tertiary structures that allows for the great functional diversity of RNA.
In her second talk, Pyle focuses on the self-splicing Group II introns. These molecules are very large ribozymes that catalyze their own splicing and transposition, employing a reaction and an active-site similar to that of the eukaryotic spliceosome. To better understand the chemistry of pre-mRNA splicincg, Pyle and her group obtained a high-resolution crystal structure of the Oceanobacillus iheyensis Group IIC intron. The crystal structure provided insights into the key roles that divalent and monovalent ions play in RNA chemistry and tertiary architecture.
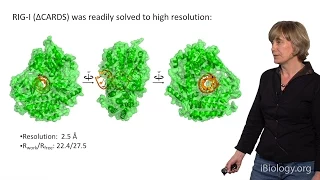
During the final talk in this series, Pyle switches her focus to a specialized class of mechanical proteins that bind and manipulate RNA molecules. This protein family includes RNA helicases, which translocate along RNA strands and strip away associated macromolecules. However, a related set of proteins display functions different from helicase activity, including a role as RNA-activated biosensors. Through crystallographic, biochemical and cell-based studies of innate immune receptor RIG-I, Pyle has shown that this human surveillance protein recognizes and binds to small viral double stranded RNAs. The subsequent binding of ATP induces protein conformational changes that contribute to signal transduction and activation of the interferon response in vivo.
Speaker Bio:
Anna Marie Pyle is the William Edward Gilbert Professor of Molecular, Cellular and Developmental Biology and Professor of Chemistry at Yale University and an Investigator of the Howard Hughes Medical Institute. Pyle received her BA from Princeton University and her PhD in Chemistry from Columbia University. She was a post-doctoral fellow with Tom Cech at the University of Colorado. Before joining Yale, Pyle was a faculty member at Columbia from 1992-2002.
Pyle’s lab uses enzymatic and biophysical methods to explore the complex structures of large RNA molecules, such as self-splicing introns. Her lab also studies the molecular motor proteins that operate on RNA, such as RNA helicases and RNA-activated biosensors that contribute to the vertebrate antiviral response.
More information is available on Dr. Pyle’s lab page at http://pylelab.org/
Lecture Overview:
In Part 1, Dr. Pyle explains that many RNA molecules have elaborate structures that are essential for their functions. Even mRNA, a relatively linear molecule, can contain distinctive three- dimensional structures. RNA duplexes are the units of secondary structure, and these form in regions where base-pairing occurs. Duplex regions often include internal or terminal loops, and they can contain unusual types of base-pairing. These secondary structural elements can arrange themselves to form highly complex tertiary structures. It is the variety of these tertiary structures that allows for the great functional diversity of RNA.
In her second talk, Pyle focuses on the self-splicing Group II introns. These molecules are very large ribozymes that catalyze their own splicing and transposition, employing a reaction and an active-site similar to that of the eukaryotic spliceosome. To better understand the chemistry of pre-mRNA splicincg, Pyle and her group obtained a high-resolution crystal structure of the Oceanobacillus iheyensis Group IIC intron. The crystal structure provided insights into the key roles that divalent and monovalent ions play in RNA chemistry and tertiary architecture.
During the final talk in this series, Pyle switches her focus to a specialized class of mechanical proteins that bind and manipulate RNA molecules. This protein family includes RNA helicases, which translocate along RNA strands and strip away associated macromolecules. However, a related set of proteins display functions different from helicase activity, including a role as RNA-activated biosensors. Through crystallographic, biochemical and cell-based studies of innate immune receptor RIG-I, Pyle has shown that this human surveillance protein recognizes and binds to small viral double stranded RNAs. The subsequent binding of ATP induces protein conformational changes that contribute to signal transduction and activation of the interferon response in vivo.
Speaker Bio:
Anna Marie Pyle is the William Edward Gilbert Professor of Molecular, Cellular and Developmental Biology and Professor of Chemistry at Yale University and an Investigator of the Howard Hughes Medical Institute. Pyle received her BA from Princeton University and her PhD in Chemistry from Columbia University. She was a post-doctoral fellow with Tom Cech at the University of Colorado. Before joining Yale, Pyle was a faculty member at Columbia from 1992-2002.
Pyle’s lab uses enzymatic and biophysical methods to explore the complex structures of large RNA molecules, such as self-splicing introns. Her lab also studies the molecular motor proteins that operate on RNA, such as RNA helicases and RNA-activated biosensors that contribute to the vertebrate antiviral response.
More information is available on Dr. Pyle’s lab page at http://pylelab.org/
- 1 participant
- 32 minutes