25 Sep 2023
Planning for Hacktoberfest project (annotating embryo data). What are the number of cell types in a human body? Cell size/function scaling and type distributions via meta-analysis. Dynamic morphogenesis and morphogenetic flows: reaction-diffusion, discrete computation, and circulation as a means to break and reinforce symmetry. Attendees: Sushmanth Reddy Mereddy, Bradly Alicea, Jesse Parent, Susan Crawford-Young, and Richard Gordon.
- 4 participants
- 1:54 hours

11 Sep 2023
DevoSAM with prompts and masks. MultiGPU cell segmentation. Overview of 2-D and 3-D Gastruloid biology and suitable computational modeling. Artificial models of cell differentiation, symmetry-breaking, and cellular decision-making. Cyanobacterial filaments as models of differential single cell morphogenesis. Attendees: Sushmanth Reddy Mereddy, Luke Knoble, Bradly Alicea, Jesse Parent, Susan Crawford-Young, and Richard Gordon.
- 4 participants
- 1:34 hours

29 Aug 2023
DevoLearn updates. DevoSAM, zero-shot learning from embryo microscopy, DevTOGL, graph embeddings. Overview of the Molecular Biology of Differentiation paper. Genetic Mosaics in Drosophila paper (from 1972) and how to make genetic mosaics. Gene Regulatory Networks of gut and intestinal differentiation in C. elegans. Attendees: Sushmanth Reddy Mereddy, Himanshu Chougule, Bradly Alicea, Jesse Parent, Morgan Hough, Lukas, and Richard Gordon.
- 5 participants
- 1:37 hours

21 Aug 2023
DevoLearn updates. How to create effective open-source documentation. CompuCell 3D workshop. Synthetic Biology circuits in cells. Temporal transcriptomics and transcriptional dynamics during the process of differentiation. How to infer cell mechanics parameters from computational models. How to infer symmetry from droplets. Attendees: Sushmanth Reddy Mereddy, Bradly Alicea, Susan Crawford-Young, Lukas, and Richard Gordon.
- 4 participants
- 1:46 hours

14 Aug 2023
DevoLearn/DevoGraph updates. Topological analysis pipeline and persistence diagrams from static data. Resources for synthetic biology and minimal cell evolution (evolving from a minimal state). A theory of molecular order? The central dogma and theories of the cell. The steps towards differentiation, understanding asymmetry in lineage trees, and cell-cell interactions. Attendees: Sushmanth Reddy Mereddy, Bradly Alicea, Susan Crawford-Young, Jesse Parent, Richard Gordon, and Himanshu Chougule.
- 4 participants
- 2:01 hours

31 Jul 2023
DevoLearn updates. Challenges of modeling in COMSOL. Genomics of Differentiation: proteomics of development, bioinformatics, comparative biology, and paper planning. Genotype to Phenotype (G-P) maps: theory, computational models, and evolution. Attendees: Sushmanth Reddy Mereddy, Bradly Alicea, Morgan Hough, Susan Crawford-Young, Richard Gordon, and Lukas.
- 5 participants
- 1:59 hours

24 Jul 2023
GSoC updates (DevoLearn and DevoGraph). MedSAM, Persistent Homology, and TOGL. Genomics of Differentiation: protein alignments for C. elegans, Global vs. Mosaic Methylation and Control of Cell Differentiation. Differentiation codes and associated analysis. Attendees: Sushmanth Reddy Mereddy, Himanshu Chougule, Bradly Alicea, Susan Crawford-Young, Richard Gordon, Jyothi Swaroop, and Lukas.
- 6 participants
- 1:49 hours

17 Jul 2023
GSoC updates, Developmental ML/DL and improving DevoLearn. Differentiation Genomics, Cell Fates, and Epigenetic Switches. June 2023 OpenWorm Newsletter. New directions in constructive connectomics. Attendees: Sushmanth Reddy Mereddy, Bradly Alicea, Susan Crawford-Young, Richard Gordon, Jyothi Swaroop, Lukas, Sai Harshitha, and Jesse Parent.
- 7 participants
- 2:02 hours

10 Jul 2023
GSoC updates (nucleus segmenter pre-topological data, DevoLearn). Open problem: genomic signatures of differentiation trees. Protein space navigation, biology is more theoretical than physics, Thompsonian grids and connections to development. Papers on directed graphs, transformers, and topological data analysis. Attendees: Sushmanth Reddy Mereddy, Bradly Alicea, Morgan Hough, Himanshu Chougule, Susan Crawford-Young, Richard Gordon, and Jesse Parent.
- 5 participants
- 1:46 hours

4 Jul 2023
GSoC Updates: Devonet and model selection, topological cell segmentation. Discussion on Topological Data Analysis (TDA) and GNNs. Large Language Models for genomics and HyenaDNA. Discussion on biomedical image sampling problem and creating biophysics graphs. Embryoids III (more on human embryoids). Attendees: Sushmanth Reddy Mereddy, Jiahang Li, Bradly Alicea, Himanshu Chougule, Susan Crawford-Young, Richard Gordon, and Jesse Parent.
- 6 participants
- 1:51 hours

26 Jun 2023
GSoC Updates: SAM and MedSAM, integrating SAM into 2-D and 3-D models of the embryo. More developments in biological embryo models (embryoids in Monkeys). Shear jamming and nonlinear phase transitions in soft biological materials (embryos and cell collectives). Attendees: Sushmanth Reddy Mereddy, Bradly Alicea, Susan Crawford-Young, Morgan Hough, and Jesse Parent.
- 3 participants
- 1:02 hours

19 Jun 2023
GSoC Updates: Segmenting cells for Topological Data Analysis, DevoLearn maintenance, DeepLabv3 vs SAM models. Recent developments in biological embryo models (beyond cell reprogramming and embryoids). Presentation on persistent homology, lineage trees, and connective anatomy. Attendees: Sushmanth Reddy Mereddy, Jiahang Li, Himanshu Chougule, Bradly Alicea, and Jesse Parent.
- 5 participants
- 1:34 hours

12 Jun 2023
GSoC Updates (devolearn @ onrender.com, SAM model manuscript, L1 phase C. elegans training data, and FR-CNNs). Overview of SynBio 2023. Expansion microscopy of connectomes using a synchrotron. Follow-up on the Gray-Scott model, chemical pattern formation (autocatalysis), and collective behavior as discrete n-wise interactions (automata, graphs). Attendees: Sushmanth Reddy Mereddy, Jyothi Swaroop, Susan Crawford-Young, Himanshu Chougule, Bradly Alicea, and Richard Gordon.
- 5 participants
- 1:43 hours
6 Jun 2023
GSoC Updates. Reaction-Diffusion Morphogenesis in 2-D and 3-D (Turing and Gray-Scott Models). Nonlinear oscillators and B-Z reactions. Connecting PDEs to C-RNNs, Cellular Automata to Reinforcement Learning. Game of Life and Particle Lenia -- Hamiltonian and Continuous representations. Attendees: Sushmanth Reddy Mereddy, Morgan Hough, Susan Crawford-Young, Himanshu Chougule, Bradly Alicea, and Richard Gordon.
- 5 participants
- 1:48 hours

30 May 2023
Using the Segment Anything Model to visualize 4-D cell lineages. Developmental-environment interactions and the Free Energy Principle, division planes in circular and triangular cells, digital biological twinning and hyperrealistic models of C. elegans (for understanding chemoattractive stimuli). Attendees: Sushmanth Reddy Mereddy, Morgan Hough, Susan Crawford-Young, Amanda Nelson, Bradly Alicea, Jiahang Li, and Richard Gordon.
- 5 participants
- 1:45 hours
23 May 2023
The fossil origins of Diatoms, Segment Anything Model and prompting with image masks and centroids, curvatures and planes in collective cell behaviors, guide to C. elegans developmental cell lineages and lineage trees (GSoC community resource). Attendees: Sushmanth Reddy Mereddy, Morgan Hough, Susan Crawford-Young, Bradly Alicea, Jiahang Li, and Richard Gordon.
- 4 participants
- 1:59 hours

15 May 2023
GSoC community period: DevoLearn/DW Media/DW Publications. Updates on implementing the Segment Anything Model on the Cell Tracking Challenge dataset. Prime Editing and genetic engineering with precision, the use of biological modules in guiding developmental processes. Cell cycles and transcriptional filtering. Attendees: Sushmanth Reddy Mereddy, Morgan Hough, Jesse Parent, Jyothi Swaroop Reddy, Susan Crawford-Young, Bradly Alicea, and Richard Gordon.
- 4 participants
- 1:34 hours

8 May 2023
Google Summer of Code onboarding and welcome. Updates on DevoLearn and embryo segmentation. Community period: getting familiar with OpenWorm, DevoWorm, and Orthogonal Research and Education Lab. Axolotl embryo images from the ball microscope. Assembly theory and the origins of life. Assembly demonstration with Braitenberg Vehicles at the edge of life. Attendees: Sushmanth Reddy Mereddy, Jiahang Li, Himanshu Chougule, Jesse Parent, Susan Crawford-Young, Bradly Alicea, and Richard Gordon.
- 5 participants
- 1:28 hours

1 May 2023
Segment Anything Model (SAM) and integrating multiple forms of cell segmentation into DevoLearn. Early Life, RNA World, and the Miller-Urey experiment. Spontaneous, ubiquitous Biochemistry. Design principles for Synthetic Mammalian Cells. New C. elegans papers on larval connectomics and low-dimensional developmental manifolds. New Attendees: Susan Crawford-Young, Jesse Parent, Richard Gordon, Sushmanth Reddy, Jyothi Swaroop, and Bradly Alicea
- 4 participants
- 1:53 hours

24 Apr 2023
Planning for a summer of DevoLearn/DevoGrah development (for development). Papers from the new book "Mathematical Biology of Diatoms". Synthetic Morphology and Antoni Gaudi's hanging chain model of bio-inspired tensegrity. Four papers on GNNs laying out the scope of future work in developmental biology. Attendees: Susan Crawford-Young, Jesse Parent, Jiahang Li, Himanshu Chougule, Sushmanth Reddy, Jyothi Swaroop, and Bradly Alicea
- 4 participants
- 2:03 hours

18 Apr 2023
Update on docs and web infrastructure for DevoLearn/DevoZoo. Papers from the special issue on Symmetries in Mind and Life (RSIF). Symmetry and asymmetry in early life, a new theory of early life, revisiting the role of symmetry-breaking in the embryo. Attendees: Susan Crawford-Young, Jesse Parent, Richard Gordon, and Bradly Alicea
- 3 participants
- 1:40 hours
10 Apr 2023
Update on docs and web infrastructure for DevoLearn/DevoZoo. New models for life's origins, history and usefulness of Artificial Life models. Learning on Graphs 2023 and graphical modeling. Papers on curvatures, modeling, and jamming phase transitions. Attendees: Sushmanth Reddy Mereddy, Susan Crawford-Young, Jesse Parent, and Bradly Alicea
- 3 participants
- 1:29 hours

27 Mar 2023
Update of devoworm.github.io and DevoLearn documentation. More detailed discussion of APS March meeting. Multiphysics modeling of the embryo and finite element alternatives. Articles on the potential of modern Biophysics as an interdisciplinary enterprise and morphogenetic curvatures. Attendees: Richard Gordon, Sushmanth Reddy Mereddy, Susan Crawford-Young, Morgan Hough, and Bradly Alicea
- 6 participants
- 1:34 hours
20 Mar 2023
Physical Modeling of Development updates, brief discussion of the APS meeting, instance segmentation discussion, GSoC and DevoLearn/ DevoGraph project updates (pull requests and open repositories), D-GNNs and graph schemas. Digits in a Dish, Curvatures (embryos) and Consendates (cellular compartments). Attendees: Richard Gordon, Sushmanth Reddy Mereddy, Susan Crawford-Young, Morgan Hough, and Bradly Alicea
- 6 participants
- 2:08 hours
13 Mar 2023
DevoLearn roadmap (semantic segmentation, instance segmentation, graphical representations, and source data). Implementing multiscale biological Agent-based Models (ABMs) using local vs. global criteria. Identifying butterfly wing spots using CNNs and implications for models of morphogenesis. Attendees: Richard Gordon, Sushmanth Reddy Mereddy, Jesse Parent, and Bradly Alicea
- 3 participants
- 1:34 hours

6 Mar 2023
Semantic Segmentation vs. Instance Segmentation for embryo cell segmentation. Update on how to propose a GSoC project. Origins of the animal embryo: phylogeny and cell biology. Alternative histories of science. Swarms, microswimmers, and chemical gradient learning. Attendees: Richard Gordon, Sushmanth Reddy Mereddy, Kritika Verma, Shalini Vaggu, and Bradly Alicea
- 5 participants
- 1:51 hours

28 Feb 2023
Progress on and getting ready for Google Summer of Code. Articles on the History of the Second Law of Thermodynamics. Biological systems as open systems (and open systems vs. closed systems). Living with Waddington (epigenetic) Landscapes, The tempo of developmental time in the genotype and phenotype. Attendees: Richard Gordon, Susan Crawford-Young, Sushmanth Reddy Mereddy, Kritika Verma, Jesse Parent, and Bradly Alicea
- 6 participants
- 1:22 hours
20 Feb 2023
Progress on DevoLearn/DevoGraph and reviewing Huggingface and Github. Multicolored embryo segmentation maps. Paper highlighting the proliferation of Mechanosensation-competent cells vs. Apoptotic cells in morphogenesis. Discussion on incommensurability (Thomas Kuhn) in different fields of biological science. Paper on applying GNNs (Graph Neural Networks) to movement of C. elegans adult movement patterns and stability. How a growing phenotype (blades of grass, stalk of wheat) maintain upright growth through coupled positive feedbacks. Paper and discussion on universality, rank-order trees and networks, and the hierarchy/heterarchy distinction. Attendees: Richard Gordon, Susan Crawford-Young, Sushmanth Reddy Mereddy, Gautham Krishnan, and Bradly Alicea
- 5 participants
- 1:30 hours

13 Feb 2023
DevoLearn/DevoWormML on Huggingface and Github. New models and repositories, open data on Huggingface. C. elegans cell modeling and cell geometry. Models of cell-cell interaction and cytoskeletons in development. Models of tensegrity networks, biological material soft physics, and network organization Mathematical modeling of spontaneous autocatalysis in vesicles: adding a toy model of Braitenberg Vehicles, a mixed molecular world, and memory mechanism for spontaneous autocatalysis. Attendees: Richard Gordon, Susan Crawford-Young, Sushmanth Reddy Mereddy, Gautham Krishnan, and Bradly Alicea
- 5 participants
- 1:25 hours
6 Feb 2023
DevoLearn/DevoWormML on Huggingface. Mathematical modeling of spontaneous autocatalysis in vesicles. Long-range interactions in morphogenesis and embryo networks, interactions within collectives of cells and extracellular matrix. Attendees: Richard Gordon, Susan Crawford-Young, Sushmanth Reddy Mereddy, Harikrishna Pillai, Gautham Krishnan, and Bradly Alicea
- 6 participants
- 1:46 hours
30 Jan 2023
Cellular packings and tensegrity networks, new open-source issues for DevoLearn and DevoGraph, Markov blankets and cell membranes, origins of early life and life from vesicles. Origins of sexual reproduction as cellular integration. How long do we have to wait for life? Attendees: Richard Gordon, Susan Crawford-Young, Sushmanth Reddy Mereddy, Barry Robson, and Bradly Alicea
- 4 participants
- 1:43 hours

23 Jan 2023
A presentation on 3-D CNNs and QCANet (segmentation for GNNs). A short discussion of MeMoDevo symposium and accepted paper in special issue of Royal Society Interface Focus on Symmetries in Biological Systems. Presentation on Biological Tensegrity Networks, with focus on static and dynamic mechanics. Attendees: Richard Gordon, Susan Crawford-Young, Sushmanth Reddy Mereddy, and Bradly Alicea
- 4 participants
- 1:49 hours
9 Jan 2023
Presentation on biological tensegrity network modeling. Discussion of Google Summer of Code 2023 project (DevoGraph and D-GNNs), Synthetic Daisies blogpost on Learning on Graphs. Paper on self-organizing biological materials and the theormodynamics of biophysics Attendees: Richard Gordon, Susan Crawford-Young, Sushmanth Reddy Mereddy, and Bradly Alicea
- 4 participants
- 1:27 hours

19 Dec 2022
Recap of the Morphing matter, Learning on Graphs, and MeMoDevo workshops, Mayukh Deb's Gradient article, Sodaconstructor (open-source Sodaplay platform) as a basis for tensegrity networks. Synthetic Biology as a testing ground for Development and Morphogenesis. Papers on a viscoplastic lock for progressive body-axis elongation and tugs-of-war between filament treadmilling and myosin-induced contractility. Attendees: Richard Gordon, Susan Crawford-Young, and Bradly Alicea
- 2 participants
- 1:44 hours

5 Dec 2022
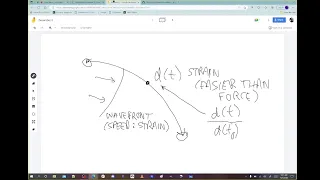
Revisiting the spherical embryo mapping (surface phenomena, from 2-D to 3-D projection). Paper on flat embryo maps (3-D to 2-D projections). Speed-Strain curves from 3-D modeling. Reaction-diffusion in 2-D vs. 3-D. Paper on the hourglass model for understanding gastrulation dynamics in mouse and rabbit. Attendees: Richard Gordon, Susan Crawford-Young, Bradly Alicea, and Morgan Hough
- 3 participants
- 1:27 hours
28 Nov 2022
Modeling tensegrity networks in MATLAB/SciLab and COMSOL. From a 3-D structure to a connectivity matrix. Components of biological tensegrity and structural propagation. Neural operators and solving for reaction-diffusion morphogenesis. Attendees: Richard Gordon, Susan Crawford-Young, Jesse Parent, Bradly Alicea, and Morgan Hough
- 4 participants
- 1:23 hours
21 Nov 2022
Creating a statistical mask from distributions. Modeling the origins of life with Braitenberg Vehicles. Mathematical modeling of autocatalytic sets for innovation and origins. Minimal cells and simulating whole bacterial cells, putting cellular components together into a whole. Attendees: Richard Gordon, Morgan Hough, Susan Crawford-Young, Jesse Parent, Bradly Alicea, and Morgan Hough
- 3 participants
- 1:17 hours

14 Nov 2022
Modeling tensegrity using COMSOL, MATLAB, and cells. DevoWorm annual activity review (for the OpenWorm annual meeting). Discussion of paper on morphogenetic waves (vs differentiation waves), and assorted papers on materials and morphogenesis. Attendees: Richard Gordon, Susan Crawford-Young, Bradly Alicea, and Morgan Hough
- 3 participants
- 1:33 hours

31 Oct 2022
Discussion of breast cancer physics, simulating tensegrity and its consequences for biological organization (left-right asymmetry and robustness). Composing Braitenberg Vehicles in development and evolution. Going from a parts list to a phenotype that works. Paper on the interactions between organismal physics (bending forces) and molecular mechanisms. Attendees: Richard Gordon, Susan Crawford-Young, Bradly Alicea, and Morgan Hough
- 4 participants
- 1:13 hours

24 Oct 2022
Simulating Lamarckism vs. Learning vs. Darwinian evolution and its connection to the acquisition of traits. Repeatability of developmental shape and tissue differentiation. The physics and shape of cells in disorders and cell populations. Self-organization of division rings in minimal cells (synthetic biology). Lightfield imaging. Attendees: Richard Gordon, Susan Crawford-Young, Bradly Alicea, and Jesse Parent
- 3 participants
- 1:18 hours

17 Oct 2022
What's new in C. elegans: multimodal integration and its connection to mammalian forms of integration, push-pull morphogenesis in the developing gonad, shape changes and stem cell differentiation, update on Bacillaria image processing. Attendees: Richard Gordon, Bradly Alicea, Morgan Hough, Susan Crawford-Young, and Alon Samuel
- 4 participants
- 1:25 hours

10 Oct 2022
Diatom dynamics and image processing, biological tensegrity in tissues and ways to assess stress and strain in embryos. Self-assembly and physical forces in cells/tissues. Formation of nervous system structures (structurogenesis) in mouse and human-derived organoids. Attendees: Bradly Alicea, Richard Gordon, Susan Crawford-Young, and Alon Samuel
- 4 participants
- 1:41 hours

3 Oct 2022
Talk on Differentiation Waves and Janus-faced logic. Discussion of mitochondrial networks, the energetic dynamics of cells, and Eukaryotic symbiosis. Attendees: Bradly Alicea, Richard Gordon, Susan Crawford-Young, and Morgan Hough
- 3 participants
- 1:20 hours

26 Sep 2022
Project updates for Cancer Imaging, what's next for DevoLearn. Neuromatch 5 preview. Components of C. elegans Connectome (synapses and neuronal diversity). Connections to tensegrity and mitochondrial energy balance. Attendees: Bradly Alicea, Richard Gordon, Susan Crawford-Young, Jesse Parent, and Harikrishna Pillai
- 3 participants
- 1:05 hours

19 Sep 2022
Discussion of mathematical modeling for Diatom movement tracking. Master plan for incorporating 2022 GSoC contributions into DevoLearn and DevoWormAI. A short tutorial on Delay Differential Equations (DDE). Papers on C. elegans transgenerational inheritance and gonad morphogenesis. Attendees: Alon Samuel, Bradly Alicea, Susan Crawford-Young, Morgan Hough, Anant Kumar, and Karan Lohaan
- 2 participants
- 1:16 hours

12 Sep 2022
Open Source contributions. Symmetry-breaking as demonstrated by cell motility and biophysical dynamics. Analysis and simulation of Drosophila eye development with high-resolution cell segmentation and EYEvolve. Attendees: Alon Samuel, Richard Gordon, Bradly Alicea, Susan Crawford-Young, Morgan Hough, Anant Kumar, and Karan Lohaan
- 4 participants
- 1:20 hours

9 Sep 2022
Contributions to Digital Bacillaria and progress on Digital Microspheres. Cell types and next-gen sequencing, developmental and regeneration in Axolotl, amniotes, and organoids. Attendees: Alon Samuel, Richard Gordon, Bradly Alicea, Susan Crawford-Young, Morgan Hough, Harikrishna Pillai, and Karan Lohaan
- 4 participants
- 1:18 hours

5 Sep 2022
Contributions to Digital Bacillaria and progress on Digital Microspheres. Cell types and next-gen sequencing, developmental and regeneration in Axolotl, amniotes, and organoids. Attendees: Alon Samuel, Richard Gordon, Bradly Alicea, Susan Crawford-Young, Morgan Hough, Harikrishna Pillai, and Karan Lohaan
- 5 participants
- 1:21 hours

29 Aug 2022
Discussion on Diatom cell tracking, physical movement mechanisms, and interspecific diversity. Push-pull Morphogenesis III: bioprinting optimization and a systems biology for development. GsoC project updates, Attendees: Alon Samuel, Richard Gordon, Bradly Alicea, Susan Crawford-Young, Morgan Hough, Harikrishna Pillai, Wataru Kawakami, and Sushmanth Mereddy
- 8 participants
- 1:20 hours
22 Aug 2022
GSoC Updates, image transformations (inferring locations on the surface of a sphere). Paper on particle tracking algorithms and traction force microscopy. D'arcy Thompson's phenotypic warping and an insect egg dataset. 3-D printed embryos and the challenges of microresolution. Attendees: Alon Samuel, Richard Gordon, Susan Crawford-Young, Bradly Alicea, Morgan Hough, Jesse Parent, Karan Lohaan, Harikrishna Pillai, Longhui Jiang
- 5 participants
- 1:28 hours

15 Aug 2022
GSoC Coding period (week 9 and 10): Updates on Digital Microspheres and D-GNNs. Review of activity on the Devolearn Github organization. A review of computing Diatoms (Digital Bacillaria). Overview of NetSci 2022. Review of the transcriptional program for regeneration in Stentor. Attendees: Richard Gordon, Jiahang Li, Morgan Hough, Alon Samuel, Joshua Brewster, and Bradly Alicea
- 5 participants
- 1:26 hours

1 Aug 2022
GSoC Coding period (week 8): Updates on Digital Microspheres and D-GNNs. Developmental Symmetries and Asymmetries. An overview of NeuroMechFly (virtual Drosophila). Discussions of applying GNNs/topological models to spatiotemporal representations of development and organisms across the tree of life. Attendees: Richard Gordon, Harikrishna Pillai, Jesse Parent, Karan Lohaan, Sushmanth Mereddy, Longhui Jiang, Anant Kumar, Jiahang Li, and Bradly Alicea
- 4 participants
- 1:13 hours

25 Jul 2022
GSoC Coding period (week 7): Updates on Digital Microspheres and D-GNNs. Mathematics of DevoWorm poster. Discussions of papers on unifying methods in Systems Biology, and a second set of papers on the physics of pattern formation (push-pull dynamics) in chick embryo and cell culture. Attendees: Susan Crawford-Young, Harikrishna Pillai, Longhui Jiang, Karan Lohaan, Morgan Hough, and Bradly Alicea
- 4 participants
- 1:16 hours
20 Jul 2022
GSoC Coding period (weeks 5 and 6): Updates on Digital Microspheres and D-GNNs. Discussions of papers on the effects of light exposure in Axolotl and chick embryos, and the physics of pattern formation (push-pull dynamics, liquid crystals) in chick and starfish embryo models. Attendees: Susan Crawford-Young, Harikrishna Pillai, Karan Lohaan, and Bradly Alicea
- 3 participants
- 1:16 hours
11 Jul 2022
GSoC Coding period (weeks 3 and 4): Updates on Digital Microspheres and D-GNNs. Activity Review (Github Issues, project tracker). Discussion about organoid modeling and differentiation trees, waves, and hypernetworks. Small-world networks as epithelial cell populations. Attendees: Susan Crawford-Young, Harikrishna Pillai, Morgan Hough, Karan Lohaan, Longhui Jiang, Jiahang Li, Wataru Kamakari, and Bradly Alicea
- 9 participants
- 1:11 hours

27 Jun 2022
GSoC Coding period (week 2): Updates on Digital Microspheres. Activity Review (Paper/Conference Submissions Document, Current and Inert Projects). Discussion about differentiation trees in the context of mosaic and regulative morphogenesis. Attendees: Susan Crawford-Young, Harikrishna Pillai, Karan Lohaan, Richard Gordon, Anlan Sun, and Bradly Alicea
- 11 participants
- 1:26 hours

20 Jun 2022
GSoC Coding period (week 1): Updates on D-GNNs and Digital Microspheres. Modeling cell and tissue tensegrity in COMSOL, review of Digital Bacillaria project and modeling movement smoothness, Papers on C. elegans positional mapping and lineage tree (embryogenetic) variation across Nematode taxonomy. Discussion about differentiation waves in the context of mosaic and regulative morphogenesis. Attendees: Susan Crawford-Young, Harikrishna Pillai, Karan Lohaan, Richard Gordon, Wataru Kawakami, and Bradly Alicea
- 11 participants
- 1:22 hours

14 Jun 2022
GSoC Community Period (week 3), getting ready for projects. Updates on D-GNNs and Digital Microspheres. Discussion on how to capture your insights and roadmap projections during GSoC (Moon shots, Mars shots, Intergalactic shots). Review of tools and education for biological and cell modeling using cellular automata and other techniques. Attendees: Susan
Crawford-Young, Jesse Parent, Harikrishna Pillai, Karan Lohaan, Richard Gordon, Jiahang Li, and Bradly Alicea
Crawford-Young, Jesse Parent, Harikrishna Pillai, Karan Lohaan, Richard Gordon, Jiahang Li, and Bradly Alicea
- 6 participants
- 1:03 hours
6 Jun 2022
GSoC Community Period (week 2) continues with some words from Tom Portegys on bio-inspired AI and robotics. GSoC intern updates. Discussion of how past modeling efforts can be used to develop a model of regenerating networks. Discussion of neural organoids and a paper on Body Plan Identity in the evolution of development. Attendees: Susan Crawford-Young, Morgan Hough, Jesse Parent, Harikrishna Pillai, Tom Portegys, Richard Gordon, Wataru Karakami, and Bradly Alicea
- 9 participants
- 1:32 hours

30 May 2022
Beginning of the GSoC Community Period (week 1). Open Source and Open Science reading repository and a tour of OpenWorm Foundation (its history and projects on the Slack team). Source datasets for the D-GNN project and a discussion of the GNN literature. Vortices, sorting, and soft material principles in Plasmodium collectives. Attendees: Susan Crawford-Young, Karan Lohaan, Harikrishna Pillai, Jiahang Li, and Bradly Alicea
- 7 participants
- 1:24 hours

23 May 2022
Introduction to GSoC 2022: Digital Microspheres and D-GNNs. Student introductions. Community period and associated resources. Benchmarks for ball microscopy. Discussion about the state of Developmental Biology and the interface between topical inclusion and theory. Question-driven developmental vs. unified theories and theory-building. Attendees: Susan Crawford-Young, Karan Lohaan, Harikrishna Pillai, Wataru Kawakami, Richard Gordon, and Bradly Alicea
- 8 participants
- 1:26 hours
9 May 2022
Point cloud contour selection for embryo modeling, 13 open problems in theoretical biology and biomathematics, the role of Synaptopodin in cell migration and dynamic networks (towards tensegrity), and a community question on the evolution of sensory systems and associated cell biology. Attendees: Susan Crawford-Young, Karan Lohaan, Richard Gordon, and Bradly Alicea (Slack question from Jesse Parent)
- 7 participants
- 1:39 hours

2 May 2022
May 2 Update. Table of Contents for special issue on "Approaches to Developmental Networks", canola seeds inside the ball microscope. Topical reviews on energetics and biophysics in the Zebrafish embryo and experimental evolution in C. elegans.
- 1 participant
- 1:00 hours
25 Apr 2022
Multiview imaging and methods for spatiotemporal integration of microscopy images, Advances in Graph Neural Networks for embryogenesis, techniques for reconstructing cell lineages from sparse annotations and cell tracking, origins of metabolic (composomes) and neuronal (peptidergic-based) networks. Attendees: Susan Crawford-Young, Karan Lohaan, Jiahang Li, Richard Gordon, and Bradly Alicea
- 7 participants
- 1:18 hours
18 Apr 2022
Data Sources for Developmental Algorithms, GSoC Question and Answer, project updates. Jamming Phase Transitions, Unjamming, and Cell-cell Signaling. Jamming and 1-D Automata. Attendees: Susan Crawford-Young, Karan Lohaan, Jiahang Li, Richard Gordon, and Bradly Alicea
- 6 participants
- 1:31 hours

11 Apr 2022
Google Summer of Code update, New Directions in Developmental (Embryo) Networks and journal special issue, one-cell embryo physics and connections to networks, tensegrity networks, instances of jamming transitions (phase transitions) in embryogenesis. Attendees: Susan Crawford-Young, Karan Lohaan, Jiahang Li, Gopinath Balaruguman, Richard Gordon, and Bradly Alicea
- 6 participants
- 1:17 hours

4 Apr 2022
Google Summer of Code update, Poisson's ratio for cellular microfilaments, the cutting-edge of Graph Neural Networks, demo of Axolotl embryo modeling and multiple viewpoint reconstruction for reconstructing 3-D images. Attendees: Susan Crawford-Young, Karan Lohaan, Harikrishna Pillai, Gopinath Balaruguman, Harini Kumar, Richard Gordon, and Bradly Alicea
- 9 participants
- 1:10 hours
28 Mar 2022
Update on soft materials and spherical projections of the Axolotl embryo, models of aneural cognition (psychophysics) in diatoms, topological approaches to morphogenesis in development and evolution. Attendees: Susan Crawford-Young, Karan Lohaan, Harikrishna Pillai, ABD, Jiahang Li, Richard Gordon, and Bradly Alicea
- 9 participants
- 1:25 hours
21 Mar 2022
Google Summer of Code projects and application process. Soft Matter and Physics of intentional and collective behavior. Synteny (Chromosomal Organization) as Developmental Building Blocks. Attendees: Susan Crawford-Young, Karan Lohaan, Harikrishna Pillai, Ishan Shanware, ABD, Jiahang Li, Ujjwal Singh, Abhipsha Das, Anh Nguyen, and Bradly Alicea
- 9 participants
- 1:16 hours

14 Mar 2022
How to write a successful GSoC proposal (join the conversation on Neurostars and Slack), examples of spatial transcriptomics in mouse and C. elegans, general principles of emergence on technical networks and in bird flocks. Attendees: Susan Crawford-Young, Karan Lohaan, Harikrishna Pillai, Ishan Shanware, Gopinath Balamurugan, and Bradly Alicea.
- 7 participants
- 1:20 hours
7 Mar 2022
Modeling DNA topology from cytogenetics data, creating a spatial transcriptomics layer in embryo models, Axolotl modeling. Select resources for Graph Neural Networks (GNNs). Soft materials, bioengineering, human perception, and collective behaviors in cells and worms. Attendees: Susan Crawford-Young, Karan Lohaan, Richard Gordon, Harikrishna Pillai, Namjote, and Bradly Alicea.
- 10 participants
- 1:21 hours

28 Feb 2022
Updates and more information on Google Summer of Code projects, Overview of ongoing projects, journal special issue on networks, Daisyworld and rein control, niche construction, and system dynamics. Cellular information processing and regulation in development. Attendees: Susan Crawford-Young, Karan Lohaan, Jesse Parent, Richard Gordon, Jiahang Li, Ishan Shanware, Harikrishna Pillai, and Bradly Alicea.
- 8 participants
- 1:17 hours

21 Feb 2022
Assorted topics (Brain Gyrification and Evo-devo, Computational Embeddings, Towhee open-source, Special Issue on Developmental Network Structures). Different ways to model GRNs (computational, biological, inferential). Discussion about genomic datasets -- ENCODE and model organisms. Integration of genomic datasets and 4-D embryo datasets. Attendees: Karan Lohaan and Bradly Alicea.
- 4 participants
- 1:39 hours

7 Feb 2022
Follow-up on recent topics (Ising Models, Tensegrity, Molecular Modeling, Wolfram Patterns in Biological Pattern Formation). Hypergraphs for Developmental Phenotypes (cell and tissue differentiation), the Cybernetics and Population Genetics of Daisyworld and biologically-rigorous Gaia models. Attendees: Mainak Deb, Richard Gordon, Karan Lohaan, Susan Crawford-Young, Jesse Parent, and Bradly Alicea.
- 8 participants
- 1:18 hours
31 Jan 2022
Tensegrity structures in cells and tissues, tensegrity networks, image processing of sea urchin skeleton, Neural CAs and skeletal pattern formation, follow-up on protein structures and hydrophobicity, computation of fitness landscapes. Attendees: Mainak Deb, Richard Gordon, Karan Lohaan, Susan Crawford-Young, Sarkath Jain, and Bradly Alicea.
- 7 participants
- 1:21 hours

24 Jan 2022
Computer vision to analyze Archaea shape variation and diversity. Transformers and machine attention for biological datasets, analysis of crumpled paper (surfaces) and fractal analysis of folded structures. Dinosaur embryos, anatomical configuration in the egg, and semi-directed late embryo behaviors. Attendees: Mainak Deb, Richard Gordon, Karan Lohaan, Susan Crawford-Young, and Bradly Alicea.
- 8 participants
- 1:16 hours

17 Jan 2022
Life to the Stars (Tardigrades and Nematodes at 10% the speed of light). Final draft of our Google Summer of Code 2022 projects. 1-D Ising models, Wolfram patterns, and Mollusk shells. Neural control of the pattern forming secretome, Stochastic biology (Stochastic vs. Deterministic models), OpenWorm Browser in Oculus Quest (Github Issue #113). Attendees: Tom Portegys, Richard Gordon, Karan Lohaan, Valentina Perricone, and Bradly Alicea.
- 12 participants
- 1:26 hours

10 Jan 2022
Looking forward into 2022. Lab preprint review on Game Theory in Development and the DevoLearn platform. Google Summer of Code 2022 projects. Papers on 3-D microscopy techniques (PIV and OCM) and the biophysics of soft material thermodynamics. Attendees: Susan Crawford-Young, Richard Gordon, Karan Lohaan, and Bradly Alicea.
- 5 participants
- 1:20 hours

14 Dec 2021
Presentation on Soft Active Materials and discussion of Liquid Crystal Biology. Discussion of Biological Intelligence and Biological Simulations in 3-D, 4-D. Recap of selected readings from NeurIPS 2021. Attendees: Susan Crawford-Young, Richard Gordon, Karan Lohaan, Valentina Perricone, and Bradly Alicea.
- 10 participants
- 1:15 hours

6 Dec 2021
Presentation on Image Processing of Microscopy (Axolotl) and Cell Tracking (Zebrafish) data. Patterning in Pennate Diatoms. Optimization of Lineage trees and spatial lineage differentiation in eutelic organisms. Cambrian embryos and the emergence of metazoan body plans. Attendees: Susan Crawford-Young, Richard Gordon, Karan Lohaan, and Bradly Alicea.
- 10 participants
- 1:23 hours

22 Nov 2021
Presentation on tissue timelines (multiple scales of temporal effects) and the role of physical stresses in measurement. Additional data analysis of Bacillaria colony velocity and acceleration. Presentation on genetic switches in synthetic biology. Papers on genetic mutations across cell lines and with respect to the germline and embryonic development. Attendees: Susan Crawford-Young, Richard Gordon, Karan Lohaan, and Bradly Alicea.
- 10 participants
- 1:19 hours

15 Nov 2021
Cursory data analysis of Bacillaria colony velocity vs. acceleration. Discussion of how stress and physical manipulation can factor into experimental results. Updates on Google Summer of Code 2022 and DevoLearn preprint. Embryogenesis animations. Papers on Turing Morphogenesis, Tissue Biomechanics, and the many facets of C. elegans Commectomics. Attendees: Susan Crawford-Young, Shruti Raj Vansh Singh, and Bradly Alicea.
- 4 participants
- 1:21 hours
9 Nov 2021
Diatom oscillations and modeling movement and upsampling microscopy images. Cellular Automata and Developmental Complexity, and papers on movement as a driver of morphogenesis. Attendees: Susan Crawford-Young, Richard Gordon, and Bradly Alicea.
- 9 participants
- 1:10 hours

1 Nov 2021
Update on submissions document and Diatom Deep Learning. Research on Diatom Model for Directional Movement and Jerkiness. Papers on Turing Morphogenesis, Branching Morphogenesis, and Soft Material analogies for Cell migration during Morphogenesis, and Attendees: Jesse Parent, Susan Crawford-Young, Richard Gordon, and Bradly Alicea.
- 10 participants
- 1:18 hours
25 Oct 2021
Overview of progress on segmenting Diatom colonies with pix2pix algorithm. Topological Data Analysis and associated methods, upcoming conferences, overviews, and Github issues. New paper on the C. elegans connectome (sexual differentiation, predation). Aspects of morphogenesis: mosaic (programmatic) vs. regulative (self-organization) development, new perspectives on Turing R-D, genetic manipulation of positional information. Attendees: Asmit Singh, R. Tharun Gowda, Karan Lohaan, Mainak Deb, Susan Crawford-Young, Richard Gordon, and Bradly Alicea.
- 8 participants
- 1:20 hours

18 Oct 2021
Overview of progress on Hacktoberfest. Regeneration and the Planarian body plan network, theoretical types of developmental networks. NASA's open-science initiative, professional development discussion, and Virtual Developmental Biology. Eric Davidson's Regulatory Genome and its computational implementations (towards a computational version of the GAL4/UAS system). Attendees: Susan Crawford-Young, Jesse Parent, and Bradly Alicea.
- 4 participants
- 1:19 hours

11 Oct 2021
Overview of progress on Diatom Deep Learning using pix2pix. Hacktoberfest reminder. Call for involvement in NMC 4.0. Deep Sets and Neural Processes. Papers on the Neurotransmitter Connectome of C. elegans and Kuramoto oscillators on a sphere. Attendees: Bradly Alicea, Ujjwal Singh, R Tharun Gowda, and Krishna Katyal.
- 5 participants
- 1:20 hours
5 Oct 2021
Hacktoberfest: how to participate, 10 years of brain emulation with OpenWorm. Papers and animations on Zebrafish embyros, Octopus aquaculture, and diverse of prehistoric marine invertebrates. Attendees: Bradly Alicea, Jesse Parent, and Susan Crawford-Young.
- 3 participants
- 1:15 hours
27 Sep 2021
Hacktoberfest plans, 2021 update to the OpenWorm Foundation. Upskilling and conference participation. Wolfram's views on rulology and metamodeling. Papers on graph processing systems, developmental hypergraphs, and topological folding in Drosophila embryo/how to capture this computationally. Attendees: Bradly Alicea, Jesse Parent, Sanjay, Susan Crawford-Young, Akshay Nair, and Madhuri Srinivasan.
- 6 participants
- 1:14 hours

20 Sep 2021
Roadmapping Digital Bacillaria for further development and Hacktoberfest. Bacillaria as a digital model of motility, colonial dynamics, Diatom biomechanics, and aneural cognition. Digital models and model organisms. Planning for Neuromatch 4 Conference. Papers on a C. elegans connectome inspired robot and the projectome of the bumblebee. Attendees: Bradly Alicea, Jesse Parent, Susan Crawford-Young, and Assaf Wodeslavsky.
- 3 participants
- 1:22 hours

14 Sep 2021
Planning for Hacktoberfest and Neuromatch 4.0. Review of group meetings board and new DevoLearn contributors/pull requests/issues. Discussion of Bacillaria non-neuronal cognition paper and Collective Pattern Generators (CoPG). Papers on the mathematical modeling of the virtual worm, universal routes to explosive (phase-transitive) phenomena, and models of developmental change, phase transitions, and transformation. Attendees: Bradly Alicea, Akshay Nair, Ujjwal Singh, and Shruti Raj Vansh Singh.
- 3 participants
- 1:18 hours

30 Aug 2021
Reviewing the current state of DevoZoo, DevoLearn, and plans for Hacktoberfest. Bacillaria Aneural Psychophysics (models and simulation). Animal embryo review. Papers on brain pathway duplication as a product of genes and behavior, and wave-driven differential morphogenesis in Dictyostelium (plus parallels with Vertebrate embryogenesis).
Attendees: Bradly Alicea, Akshay Nair, and Mainak Deb.
Attendees: Bradly Alicea, Akshay Nair, and Mainak Deb.
- 4 participants
- 1:17 hours

23 Aug 2021
Considering the Future of DevoLearn, Digital Shape and Geometry in Liquid Droplets and Axolotl. Liquid Crystal Biology (LCB), Gene Expression, and Turing Morphogenesis. More on the historical contingent pathways of life, Shape, Size, and Muscular Behavior in Cnidaria. Attendees: Bradly Alicea, Shruti Raj Vansh Singh, Jesse Parent, Akshay Nair, Susan Crawford-Young, and Mainak Deb.
- 5 participants
- 1:21 hours
16 Aug 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), Size Scaling, Timing, and Division Dynamics in Single-celled contexts. Liquid Crystal Biology (LCB). Historical contingencies and replaying the tape of life, Virtual Environments for training computational developmental agents. Attendees: Bradly Alicea, Krishna Katyal, Jesse Parent, Akshay Nair, and Mainak Deb.
- 7 participants
- 1:11 hours

9 Aug 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), Review of submissions document. Semantic Versioning for Open-source Software. Game Theory of Developmental Processes preview. Developmental Landscapes for C. elegans, Liquid Crystal Biology (LCB), and Biological Symmetry-breaking. Attendees: Bradly Alicea, Krishna Katyal, Susan Crawford-Young, Akshay Nair, and Mainak Deb.
- 7 participants
- 1:18 hours
2 Aug 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), Review of submissions document and Mathematics of DevoWorm document. Immediate Early Genes and Activity-dependence in embryos and the brain, Early Embryos and the Emergence of Embryonic Development. Attendees: Bradly Alicea, Krishna Katyal, Susan Crawford-Young, Akshay Nair, and Mainak Deb.
- 6 participants
- 1:15 hours

26 Jul 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), Review of submissions document and group-wide task board. Developmental Agents, Germ Layers, and Differentiation Trees. Thermotaxis and environment in C. elegans, Cell Regulatory Networks, Flexibility of Turing Morphogenesis. Attendees: Bradly Alicea, Shruti Raj Vansh Singh, Sanjay from Amrita University, Jesse Parent, and Mainak Deb.
- 6 participants
- 1:16 hours

19 Jul 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), Review of submissions document and group-wide task board. The role of temperature in development, ecology, and evolution. Popular articles on the process of metamorphosis in insects. Papers on Cephalopod Cognition and Neuromechanical modeling in Drosophila larvae. Attendees: Akshay Nair, Bradly Alicea, Ujjwal Singh, Sanjay from Amrita University, and Mainak Deb.
- 5 participants
- 1:17 hours
12 Jul 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), review of Networks 2021 activities and ANNs/BNNs research. Review of potential tasks and group-wide task board, versioning of DevoLearn. Papers on role of light in visual and developmental environment, RoboWorm paper, and cell tracking and quantification in induced pluripotent stem cells. Attendees: Akshay Nair, Bradly Alicea, Assaf Wodeslavsky, and Mainak Deb.
- 3 participants
- 1:20 hours
28 Jun 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), Brain Networks: NetNeuro preview, brain network modeling, and parallel networks in development, the generativity of differential growth, vasculature networks, and building (thinking) scaffolds for intelligent materials. Attendees: Susan Crawford-Young, Shruti Raj Vansh Singh, Richard Gordon, Krishna Katyal, Bradly Alicea, and Mainak Deb.
- 6 participants
- 1:18 hours

21 Jun 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), memorial for Steve McGrew (holography, microscopy, genetic algorithms), discussion on scaling in the ball microscope, and papers on C. elegans Metabolic Networks (WormPaths) and Neural Progenitor migration in Octopus brain development. Attendees: Susan Crawford-Young, Richard Gordon, Bradly Alicea, and Mainak Deb.
- 4 participants
- 1:17 hours

14 Jun 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), review of Networks 2021 submissions, Katatrepsis of Insect Embryos. Papers on the natural history of model organism development (Zebra Finch), 15 years after NKS, simulating primate visual cortex using CNNs, and compound imaging techniques. Attendees: Akshay Nair, Tom Portegys, Jesse Parent, Richard Gordon, Bradly Alicea, Ujjwal Singh, Assaf Wodeslavsky, Shruti Raj Vansh Singh, and Mainak Deb.
- 5 participants
- 1:12 hours

7 Jun 2021
Update on Summer of Code activities (Mainak Deb, Improving DevoLearn), OpenWorm poster for the International C. elegans Conference, short lecture on Cell Shape Geometries in Archaea. Papers on synthetic development and embryogenesis via gastruloid. Attendees: Jesse Parent, Richard Gordon, Bradly Alicea, Shruti Raj Vansh Singh, and Mainak Deb.
- 5 participants
- 1:02 hours

31 May 2021
Review of GSoC Community Period and Open-source Community Resources, OpenWorm turns 10!, presentation on Mainak's project (Improving DevoLearn), discussion about interpolating microscopy images/data, developmental game theory preview. Paper review on information processing in cells and symmetry in development. Attendees: Jesse Parent, Richard Gordon, Susan Crawford-Young, Bradly Alicea, Shruti Raj Vansh Singh, Assaf Wodeslavsky, Akshay Nair, Ujjwal Singh, and Mainak Deb.
- 6 participants
- 1:21 hours
17 May 2021
Review of Onboarding Guide and Submissions table, discussion of network science and accepted submissions to Networks 2021 (TopoNets, NetNeuro), updates and Bacillaria research, more Paramecium non-neuronal cognition, Developmental trajectories in embryos (cricket synticium) and human brains (from embryogenesis to life-history). Attendees: R Tharun Gowda, Vrutik Rabadia, Jesse Parent, Richard Gordon, Susan Crawford-Young, Tom Portegys, Bradly Alicea, Shruti Raj Vansh Singh, Aswarth Narayama, Yash
Vadi, Akshay Nair, Ujjwal Singh, and Mainak Deb.
Vadi, Akshay Nair, Ujjwal Singh, and Mainak Deb.
- 9 participants
- 1:18 hours
10 May 2021
Presentations on Neural Cellular Automata and Regenerative Capacity (with discussion). Differentiation tree finding and regeneration, Mathematics of DevoWorm, HOX collinearity, body plans, and shape. Attendees: R Tharun Gowda, Vrutik Rabadia, Jesse Parent, Richard Gordon, Susan Crawford-Young, Krishna Katyal, Bradly Alicea, Shruti Raj Vansh Singh, Yash Vadi, and Mainak Deb.
- 10 participants
- 1:18 hours

3 May 2021
DevoLearn update and Open-source sustainability, Parallel biological networks (embryo networks and connectomes), biology of Axolotls, branching in Artificial Neural Networks, Learning and Morphogenesis in single cells, interval timing in development. Attendees: R Tharun Gowda, Jesse Parent, Richard Gordon, Susan Crawford-Young, Krishna Katyal, Bradly Alicea, Shruti Raj Vansh Singh, and Akshay Nair.
- 10 participants
- 1:21 hours
26 Apr 2021
Review of Seashell Pattern Recognition, Embryo Networks, Connectomes, and Euler Paths for Life, Digital Bacillaria review, Fish Ladder model of Abiogenesis, Optic Cups in Vertebrate Embryos and Organoids, Plant Non-neuronal Cognition, and Spatial Differentiation in Organoids. Attendees: R Tharun Gowda, Jesse Parent, Richard Gordon, Susan Crawford-Young, Krishna Katyal, Vrutik Rabadia, Bradly Alicea, Mainak Deb, Vash Yadi, Shruti Raj Vansh Singh, and Akshay Nair.
- 9 participants
- 1:17 hours

19 Apr 2021
Graph Neural Networks, Cellular Automata rule discovery using Neural Networks, Diatom Non-neuron Cognition, Neuromatch Academy, Origins of Developmental Systems and Ecological Niches. Attendees: R Tharun Gowda, Debojyoti Chakraborty, Ujjwal Singh, Jesse Parent, Richard Gordon, Susan Crawford-Young, Krishna Katyal, Vrutik Rabadia, Bradly Alicea, Mainak Deb, Vash Yadi, Abhishek Tiwari, Shruti Raj Vansh Singh, and Akshay Nair
- 11 participants
- 1:24 hours
12 Apr 2021
Separability and Overlap in Cell Microscopy, Discussion About Version-controlled Books, Correlation Does Not Equal Causation, Bioelectricity as a Form of Organismal Coherence and Cybernetic Logic as a Form of Organismal Branch Proliferation, Learning and Memory in Developmental Systems. Attendees: R Tharun Gowda, Debojyoti Chakraborty, Ujjwal Singh, Jesse Parent, Richard Gordon, Susan Crawford-Young, Krishna Katyal, Vrutik Rabadia, Bradly Alicea, Mayukh Deb, Mainak Deb, Vash Yadi, Aswath Narayana, Aayush Kumar, Shruti Raj Vansh Singh, and Muhammed Abdullah
- 14 participants
- 1:27 hours
5 Apr 2021
How to Program Cellular Automata and create rules of life, DevoWorm onboarding guide, Project board updates, Neural Organoids, wiring neural circuits II. Attendees: R Tharun Gowda, Ujjwal Singh, Jesse Parent, Richard Gordon, Susan Crawford-Young, Krishna Katyal, Bradly Alicea, Mayukh Deb, Mainak Deb, Aswath Narayana, Aayush Kumar, Shruti Raj Vansh Singh, Akshay Nair, and Muhammed Abdullah
- 9 participants
- 1:23 hours

29 Mar 2021
DevoWorm Onboarding Guide, Mathematics of DevoWorm, Project Board and Open-source Maintanence, Neural Cellular Automata, Paleontology of Development, and Wiring up Neural Circuits. Attendees: R Tharun Gowda, Abhishek Tiwari, Jesse Parent, Yash Vadi, Krishna Katyal, Bradly Alicea, Mayukh Deb, Mainak Deb, Susan Crawford-Young, Vrutik Rabadia, Zura Isakadze, and Muhammed Abdullah
- 6 participants
- 1:34 hours

22 Mar 2021
DevoWorm Onboarding Guide, Emerging GSoC-related Projects, Mathematics of DevoWorm, Superresolution, Cellular Automata and Artificial Chemistries, and Blastoids. Attendees: R Tharun Gowda, Jesse Parent, Aayush Kumar, Yash Vadi, Krishna Katyal, Bradly Alicea, Mayukh Deb, Mainak Deb, Akshay Nair, Susan Crawford-Young, Richard Gordon, Tom Portegys, Vrutik Rabadia, Debojyoti Chakraborty, and Shruti Raj Vansh Singh
- 10 participants
- 1:30 hours

15 Mar 2021
Fuzzy c-means clustering for detecting Diatom filaments, spatial modeling of the C. elegans embryo, Cellular Automata and the Game of Life for Pattern Formation, GSoC (and DevoWorm) Onboarding guide, and the modeling of emergence in embryogenesis. Attendees: R Tharun Gowda, Aayush Kumar, Yash Vadi, Krishna Katyal, Bradly Alicea, Mayukh Deb, Mainak Deb, Akshay Nair, Susan Crawford-Young, Richard Gordon, Markus Heimerl, Param Mirani, Kush Kothari, Abhishek Tiwari, and Shruti Raj Vansh Singh
- 9 participants
- 1:29 hours
8 Mar 2021
Deep Learning for Cell Division (ResNet) and Diatom Colonies (YOLO). Custom Cellular Automata for Pattern Formation, Cellular Reprogramming, and Autophagy. Ball microscopy, GSoC Onboarding, and Zebrafish goodies. Attendees: R Tharun Gowda, Assaf Wodeslavsky, Aayush Kumar, Yash Vadi, Krishna Katyal, Bradly Alicea, Mayukh Deb, Mainak Deb, Akshay Nair, Ujjwal Singh, and Shruti Raj Vansh Singh
- 9 participants
- 1:14 hours

1 Mar 2021
DevoLearn Inference Engine discussion, details of the Digital Microsphere, upcoming submissions and deadlines, Gene Regulatory Networks, Mechanotransduction and Cell Fate, and the Bioinformatics of Protein-Protein Interaction Networks. Attendees: Susan Crawford-Young, R Tharun Gowda, Bradly Alicea, Mayukh Deb, Mainak Deb, Jesse Parent, Richard Gordon, Markus Heimerl, Akshay Nair, Ujjwal Singh, and Shruti Raj Vansh Singh
- 5 participants
- 1:29 hours

22 Feb 2021
DevoLearn 0.3.0 release, Positive Feedback of Bias in AI, Voice Recognition using ResNet, the intellectual history of C. elegans neurodevelopment, the development of multicellular shape in soap bubbles, embryos, and cell colonies. Attendees: Susan Crawford-Young, R Tharun Gowda, Bradly Alicea, Krishna Katyal, Mainak Deb, Jesse Parent, and Shruti Raj Vansh Singh
- 10 participants
- 1:34 hours
15 Feb 2021
Darwin Day blog post (origins of cognition, model evo-devo-neuro systems, and phylogeography), lineage population prediction update, Ant, Spider, Centipede, and Plant Embryogenesis, and Positional Information (PI) theory. Attendees: Susan Crawford-Young, R Tharun Gowda, Bradly Alicea, Krishna Katyal, Mainak Deb, Richard Gordon, and Shruti Raj Vansh Singh
- 7 participants
- 1:18 hours

8 Feb 2021
Attendees: Advances in DevoLearn and DL Lightning Talk, Deep Learning and Art (Style Transfer using ResNet), Systems Development in Model Systems (C. elegans and Organoids). Susan Crawford-Young, Mayukh Deb, R Tharun Gowda, Bradly Alicea, Krishna Katyal, Mainak Deb, Jesse Parent, Assaf Wodeslavsky, and Shruti Raj Vansh Singh
- 4 participants
- 1:18 hours
1 Feb 2021
DevoWorm (2021, Meeting 4): ResNet training with the EPIC dataset and pre-trained model improvement, "Killing the Winner" and "Survival of the Systems", comparing trees with metrics, a billion years of biogeography, and comparisons between evolutionary algorithms and biological evolution. Attendees: Susan Crawford-Young, Bradly Alicea, Krishna Katyal, Mainak Deb, Jesse Parent, Richard Gordon, and Shruti Raj Vansh Singh
- 9 participants
- 1:21 hours

25 Jan 2021
DevoWorm (2021, Meeting 3): Issues board, Member Achievements, Submission to Evolution conference, Agent-based simulations of network diffusion, deep evolutionary history figure and the Boring Billion, papers on ML/Development and the History of Developmental Theory. Attendees: Susan Crawford-Young, Bradly Alicea, Krishna Katyal, Jesse Parent, and Shruti Raj Vansh Singh
- 3 participants
- 1:25 hours

18 Jan 2021
DevoWorm (2021, Meeting 2): Overview of New and Upcoming Publications, Finalized GSoC Projects, Growth, Form, Morphogenesis, and Machine Learning, Physics of Embryogenesis and the Molecular Milieu. Attendees: Susan Crawford-Young, Ujjwal Singh, Krishna Katyal, Shruti Rajvanshsingh, and Bradly Alicea.
- 8 participants
- 1:17 hours

11 Jan 2021
DevoWorm (2021, Meeting 1): Overview of 2020 presentation, review of caandidate GSoC projects, papers on condensates in Eukaryotic cells, Expansion Microscopy in C. elegans, and the first (Precambrian) embryos. Attendees: Susan Crawford-Young, Mainak Deb, Jesse Parent, Ujjwal Singh, Arnab Banerjee, and Bradly Alicea.
- 7 participants
- 1:21 hours
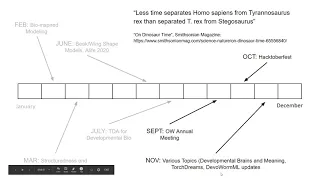
21 Dec 2020
DevoWorm (2020, Meeting 43): Papers on Physics, Embryogenesis, and Evolution. Manuscripts in the Pipeline (Boring Billion, Periodicity in the Embryo, DevoLearn), Google Summer of Code applications for 2021. Attendees: Susan Crawford-Young, Mainak Deb, Mayukh Deb, Richard Gordon, Ujjwal Singh, Shruti Rajvanshsingh, and Bradly Alicea.
- 7 participants
- 1:18 hours

14 Dec 2020
An update to the DevoWormML lecture on Input Data, "Periodicity in the Embryo" manuscript, paper on the Physics of Cell Aggregates, and Computational Biology Education. Attendees: Susan Crawford-Young, Krishna Katyal, Mayukh Deb, Mainak Deb, Jesse Parent, Debojyoti Chakraborty, and Bradly Alicea.
- 9 participants
- 1:29 hours
30 Nov 2020
Papers on Temporal Scaling and Deep Neural Control, an update to the DevoWormML lecture on Computational Pareidolia, and a review of the task board. Attendees: Susan Crawford-Young, Krishna Katyal, Shruti Rajvanshsingh, and Bradly Alicea.
- 5 participants
- 1:28 hours

23 Nov 2020
Papers on Optical Elastography and Gigapxiels, finishing up DevoLean software paper and demo of Torch Dreams, plus papers on Cell-cell Interactions and Biological Agency. Attendees: Susan Crawford-Young, Richard Gordon, Mayukh Deb, Krishna Katyal, Shruti Rajvanshsingh, Assaf Wodeslavsky, Jesse Parent, and Bradly Alicea.
- 13 participants
- 1:15 hours

16 Nov 2020
Task Review Board, discussion/presentation on SARSA reinforcement learning and general meta-feature models, and papers from the reading queue (development of brains, cell types, and meaning). Attendees: Susan Crawford-Young, Krishna Katyal, Shruti Rajvanshsingh, Jesse Parent, Bradly Alicea.
- 7 participants
- 1:26 hours
9 Nov 2020
Review of "Periodicity in the Embryo" manuscript, Notetaking Strategies (Evergreen Method, Zettelkasten), and Papers from the Reading Queue on Connectome and Cell-type Dependent Morphogenesis. Attendees: Susan Crawford-Young, Richard Gordon, Bradly Alicea.
- 5 participants
- 1:04 hours

2 Nov 2020
Recap of Neuromatch presentations, non-neuronal cognition in cell colonies, various topics in developmental systems, and papers from the reading queue. Attendees: Susan Crawford-Young, Bradly Alicea.
- 4 participants
- 1:18 hours

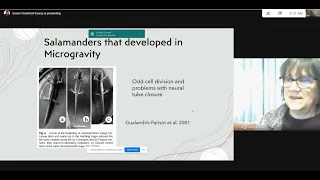
12 Oct 2020
Presentation on embryos in microgravity, review of open tasks and issues, papers from the reading queue. Attendees: Susan Crawford-Young, Bradly Alicea, Jesse Parent, Ujjwal Singh.
- 3 participants
- 1:16 hours
5 Oct 2020
Discussion on working with and encoding biological problems in software, Hacktoberfest updates, and a presentation on PCA, tSNE, and UMAP.
- 5 participants
- 1:18 hours

28 Sep 2020
Attendees: Bradly Alicea, Krishna Katyal, Susan Crawford-Young, Ujjwal Singh, and Jesse Parent. Featuring a talk by Susan titled "Optical Coherence Tomography of Live Tissue"
- 7 participants
- 1:02 hours
14 Sep 2020
Attendees: Bradly Alicea, Mayukh Deb, Mainak Deb, Ujjwal Singh, and Jesse Parent.
- 6 participants
- 60 minutes
7 Sep 2020
Attendees: Bradly Alicea, Mayukh Deb, Mainak Deb, Ujjwal Singh, and Richard Gordon.
- 5 participants
- 54 minutes

31 Aug 2020
Attendees: Bradly Alicea, Mayukh Deb, Ujjwal Singh, Maniak Deb, Susan Crawford-Young, Jesse Parent, Tom Portegys, and Richard Gordon.
- 6 participants
- 1:20 hours

24 Aug 2020
Attendees: Bradly Alicea, Mayukh Deb, Ujjwal Singh, Susan Crawford-Young, Jesse Parent, and Richard Gordon.
- 6 participants
- 1:02 hours

17 Aug 2020
Attendees: Bradly Alicea, Mayukh Deb, Ujjwal Singh, Susan Crawford-Young, Jesse Parent, and Krishna Katyal.
- 8 participants
- 1:07 hours

10 Aug 2020
Attendees: Bradly Alicea, Mayukh Deb, Ujjwal Singh, Susan Crawford-Young, Jesse Parent, and Richard Gordon.
- 9 participants
- 1:15 hours

6 Aug 2020
Attendees: Bradly Alicea, Mayukh Deb, Ujjwal Singh, Susan Crawford-Young, Jesse Parent, and Krishna Katyal. Preview of DevoLearn ML/DL platform, umbrella website, and Axolotl montaging.
- 7 participants
- 1:20 hours
27 Jul 2020
Attendees: Bradly Alicea, Mayukh Deb, Ujjwal Singh, and Richard Gordon. GSoC Updates and discussion, discussion of open project issues, papers from the reading queue.
- 5 participants
- 1:01 hours

20 Jul 2020
Attendees: Bradly Alicea, Mayukh Deb, Vinay Varma, Ujjwal Singh, Susan Crawford-Young, and Abhishek Bvs. GSoC Updates and discussion, paper planning for Periodicity of the Embryo.
- 5 participants
- 1:14 hours
13 Jul 2020
Attendees: Bradly Alicea, Mayukh Deb, Jesse Parent, Ujjwal Singh, and Krishna Katyal. GSoC Updates and discussion, paper planning for Periodicity of the Embryo.
- 6 participants
- 1:06 hours
6 Jul 2020
Attendees: Bradly Alicea, Mayukh Deb, Jesse Parent, Ujjwal Singh, and Vinay Varma. GSoC Updates, project board, and paper on Topological Data Analysis (TDA).
- 6 participants
- 1:09 hours

29 Jun 2020
Attendees: Bradly Alicea, Mayukh Deb, Jesse Parent, Ujjwal Singh, Richard Gordon and Vinay Varma.
Featuring topics such as unsupervised learning as a developmental analogue, morphological agents, and deep chicken terminator
Featuring topics such as unsupervised learning as a developmental analogue, morphological agents, and deep chicken terminator
- 10 participants
- 1:15 hours
15 Jun 2020
Attendees: Bradly Alicea, Mayukh Deb, Jesse Parent, Ujjwal Singh, Richard Gordon, Abhishek Bvs, and Vinay Varma.
- 6 participants
- 58 minutes

8 Jun 2020
Attendees: Bradly Alicea, Susan Crawford-Young, Mayukh Deb, Steve McGrew, Ujjwal Singh, Richard Gordon, and Vinay Varma.
- 7 participants
- 1:09 hours

1 Jun 2020
Attendees: Bradly Alicea, Susan Crawford-Young, Mayukh Deb, Steve McGrew, Ujjwal Singh, Richard Gordon, and Vinay Varma.
- 7 participants
- 1:11 hours

18 May 2020
DevoWorm meeting: May 18, 2020. Attendees: Richard Gordon, Jesse Parent, Mayukh Deb, Bradly Alicea, and Susan Crawford-Young.
- 9 participants
- 1:04 hours

11 May 2020
DevoWorm meeting: May 11, 2020. Attendees: Richard Gordon, Ujjwal Singh, Mayukh Deb, Krishna Katyal, Bradly Alicea, and Steve McGrew.
- 5 participants
- 1:19 hours
4 May 2020
DevoWorm meeting: May 4, 2020. Attendees: Richard Gordon, Ujjwal Singh, Bradly Alicea, Susan Crawford-Young, and Steve McGrew. Look for the "May the Fourth" easter egg at 17:47!
- 13 participants
- 1:12 hours

27 Apr 2020
DevoWorm meeting: April 27, 2020. Attendees: Richard Gordon, Ujjwal Singh, Bradly Alicea, and Jesse Parent.
- 4 participants
- 1:02 hours

20 Apr 2020
DevoWorm meeting: April 20, 2020. Attendees: Richard Gordon, Ujjwal Singh, Bradly Alicea, Vinay Varma, and Susan Crawford-Young
- 6 participants
- 57 minutes

13 Apr 2020
DevoWorm meeting: April 13, 2020. Attendees: Richard Gordon, Ujjwal Singh, Bradly Alicea, Vinay Varma, and Steve McGrew
- 8 participants
- 1:10 hours
6 Apr 2020
DevoWorm meeting: April 6, 2020. Attendees: Richard Gordon, Ujjwal Singh, Bradly Alicea, Tom Portegys, Susan Crawford-Young, Vinay Varma, and Jesse Parent
- 7 participants
- 1:05 hours

23 Mar 2020
DevoWorm meeting: March 23, 2020. Attendees: Richard Gordon, Devansh Batra, Ujjwal Singh, Bradly Alicea, Yash Patel, Susan Crawford-Young, Amrendra Pratap Singh, and Jesse Parent
- 2 participants
- 58 minutes

24 Feb 2020
DevoWorm meeting: February 24, 2020. Attendees: Richard Gordon, Vinay Varma, Devansh Batra, Thomas Portegys, Ujjwal Singh, Bradly Alicea, Yash Patel, and Jesse Parent
- 8 participants
- 1:02 hours

17 Feb 2020
DevoWorm meeting: February 17, 2020. Attendees: Richard Gordon, Vinay Varma, Susan Crawford-Young, Devansh Batra, Thomas Portegys, Emily Thomas, Ujjwal Singh, Bradly Alicea, Yash Patel, Thomas Harbich, and Jesse Parent
- 6 participants
- 60 minutes
27 Jan 2020
DevoWorm meeting: January 27, 2020. Attendees: Richard Gordon, Vinay Varma, Devansh Batra, Yash Agarwal, Bradly Alicea, and Jesse Parent
- 8 participants
- 1:02 hours
20 Jan 2020
DevoWorm meeting: January 20, 2020. Attendees: Susan Crawford-Young, Richard Gordon, Vinay Varma, Devansh Batra, Yash Agarwal, Bradly Alicea, and Jesse Parent
- 7 participants
- 60 minutes

13 Jan 2020
DevoWorm meeting: January 13, 2020. Attendees: Richard Gordon, Ujjwal Singh, Devansh Batra, Yash Agarwal, Bradly Alicea, and Jesse Parent
- 3 participants
- 1:03 hours

